1600 1800 Analysis and visualization of microbiome profile with Phyloseq. A tutorial on how to use Plotlys R graphing library for microbiome data analysis and visualization.
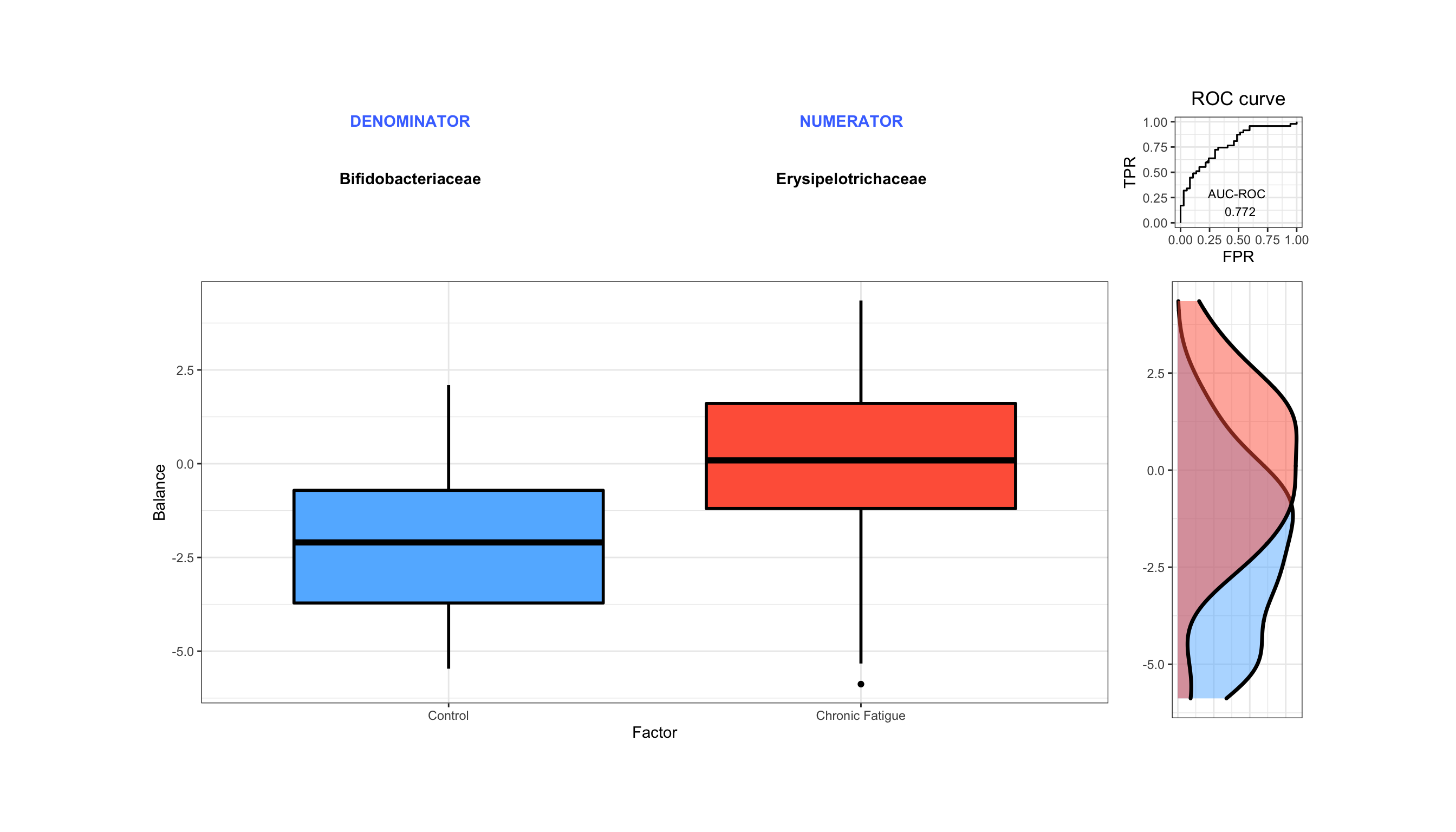
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
Normalizing count data 4.

. The microbiome R package facilitates phyloseq-based exploration and analysis of taxonomic profiling data. The data itself may originate from widely different sources such as the microbiomes of humans soils surface and ocean. Søg efter jobs der relaterer sig til Microbiome analysis in r tutorial eller ansæt på verdens største freelance-markedsplads med 21m jobs.
16S microbiome analysis of ATCC R Microbiome Standards from CJ Bioscience Inc. Please follow the below tutorial. In the microbiome network a node represents taxon and links exist between a pair of nodes if their sequence frequency are significantly correlated.
The workshop will be conducted virtually in a guided-tutorial format in which attendees will follow tutorials and instructors will guide the pace. A Hands-On Tutorial Amanda Birmingham Center for Computational Biology Bioinformatics University of California at San Diego. The purpose of this tutorial is to describe the steps required to perform.
Here we describe in detail and step by step the process of building analyzing and visualizing microbiome networks from operational taxonomic unit OTU tables in R and RStudio using several different approaches and extensively commented code snippets. The next 2 classes lessons 7 and 8 will introduce and work through the data analysis tutorial using the dada2 R package. Extending the phyloseq class.
Loading data into phyloseq 5. The sequencing reads have to be denoised and. Complete Homework 3 to be sure you are prepared to work through these exercises.
High-throughput sequencing of PCR-amplified taxonomic markers like the 16S rRNA gene has enabled a new level of analysis of complex bacterial communities known as microbiomes. Tutorial performed on a. 10 rows BEFORE YOU START.
Alternating lecture and tutorial on command-line software. Analysis of beta diversity. Therefore we created R microeco package.
During this session we will cover the fundamentals of amplicon-based microbiome analysis. This vignette provides a brief overview with example data sets from published microbiome profiling studies. The analysis of microbial communities brings many challenges.
This can be difficult for taxonomic data since. While we continue to maintain this R package the development has been. This is a short example analysis to give you a feel for how metacoder and taxa are used in microbiome analysis.
The first step in any analysis is getting your data into R. There are many great resources for conducting microbiome data analysis in R. R provides a cohesive environment to analyze data using modular toolboxes called R packages.
Multiple staticstical approaces such as pearson. With multiple example data sets from published studies. Moving microbes from the 50 percent human artscience project.
Introduction to the microbiome R package. Now we have loaded the required files we will be using those OTU count data and taxonomy file to build a microbiome network. Many tools exist to quantify and compare abundance levels or OTU composition of communities in different conditions.
Loading microbiome data into R 3. I am a microbial ecologist which means I study how microbes interact with each other and their environment. For those looking for an end-to-end workflow for amplicon data in R I highly recommend Ben Callahans F1000 Research paper Bioconductor Workflow for Microbiome.
Recently developed culture-independent methods based on high-throughput sequencing of 16S18S ribosomal RNA gene variable regions and internal transcribed spacers ITS enable researchers to identify all the microbes in their complex habitats or in other words to analyse a microbiome. An R package for microbial community analysis in an environmental context. The integration of many different types of data with methods from ecology genetics phylogenetics network analysis visualization and testing.
R version 410 2021-05-18. As a beginner the entire process from sample collection to analysis for sequencing data is a daunting task. We will do the following in this tutorial.
Fast flexible and modularized. If something does not make sense now dont worry. R6 Class to store and analyze data.
Introduction Set Up Your Environment The Microbiome Data Download and Install necessary R packages Set up Working Environment Reads The Analysis Check Read Quality Read Filtering Learn the Error Rates and Infer Sequences Merge Forward and Reverse Reads Construct Sequence Table Remove chimeras Tracking Reads throughout Pipeline Assign Taxonomy. The tutorial starts. We will cover everything shown here in greater detail later.
This is a tutorial to analyze microbiome data with R. The package is in Bioconductor and aims to provide a comprehensive collection of tools and tutorials with a particular focus on amplicon sequencing data. Tools for microbiome analysis.
R is an open source free statistical programming and graphing language that includes tools for analysis of statistical ecological diversity and community data among many other things. Explore microbiome profiles using R. Files for part 2 of the dada2 exercise at located in the Github repo at https.
Det er gratis at tilmelde sig og byde på jobs. A list of R environment based tools for microbiome data exploration statistical analysis and visualization. Statistical Analysis of Microbiome Data in R by Xia Sun and Chen 2018 is an excellent textbook in this area.
There has been a lot of R packages created for the microbiome profiling analysis. Analysis of alpha diversity 7. Workshop will use R and RStudio which are remotely accessible.
Details of the individual session components are included below. More specifically the downstream processing of raw reads is the most time consuming and mentally draining stage. Microbiome Analysis with QIIME2.
A more comprehensive tutorial is available on-line. Microbiome Analysis - dada2 Tutorial. However it is still difficult to perform data mining fast and efficiently.

Ggtree For Microbiome Data R Bloggers Data Microbiome Data Analysis
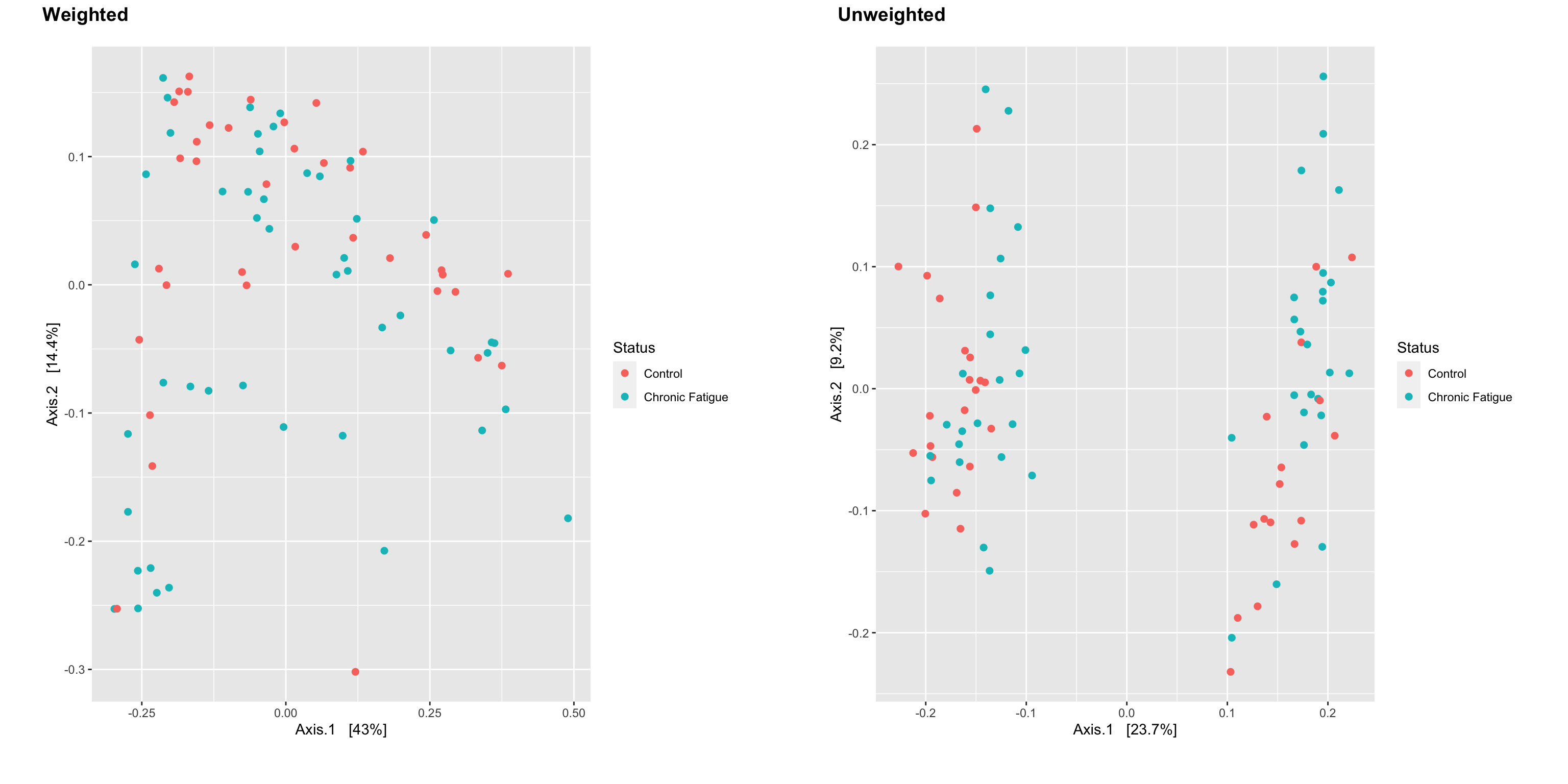
Introduction To The Statistical Analysis Of Microbiome Data In R Academic

Researchers From The Quadram Institute Have Carried Out A Meta Analysis Of The Gut Microbiome In Parkinson S Disease Gut Health Microbiome Parkinsons Disease
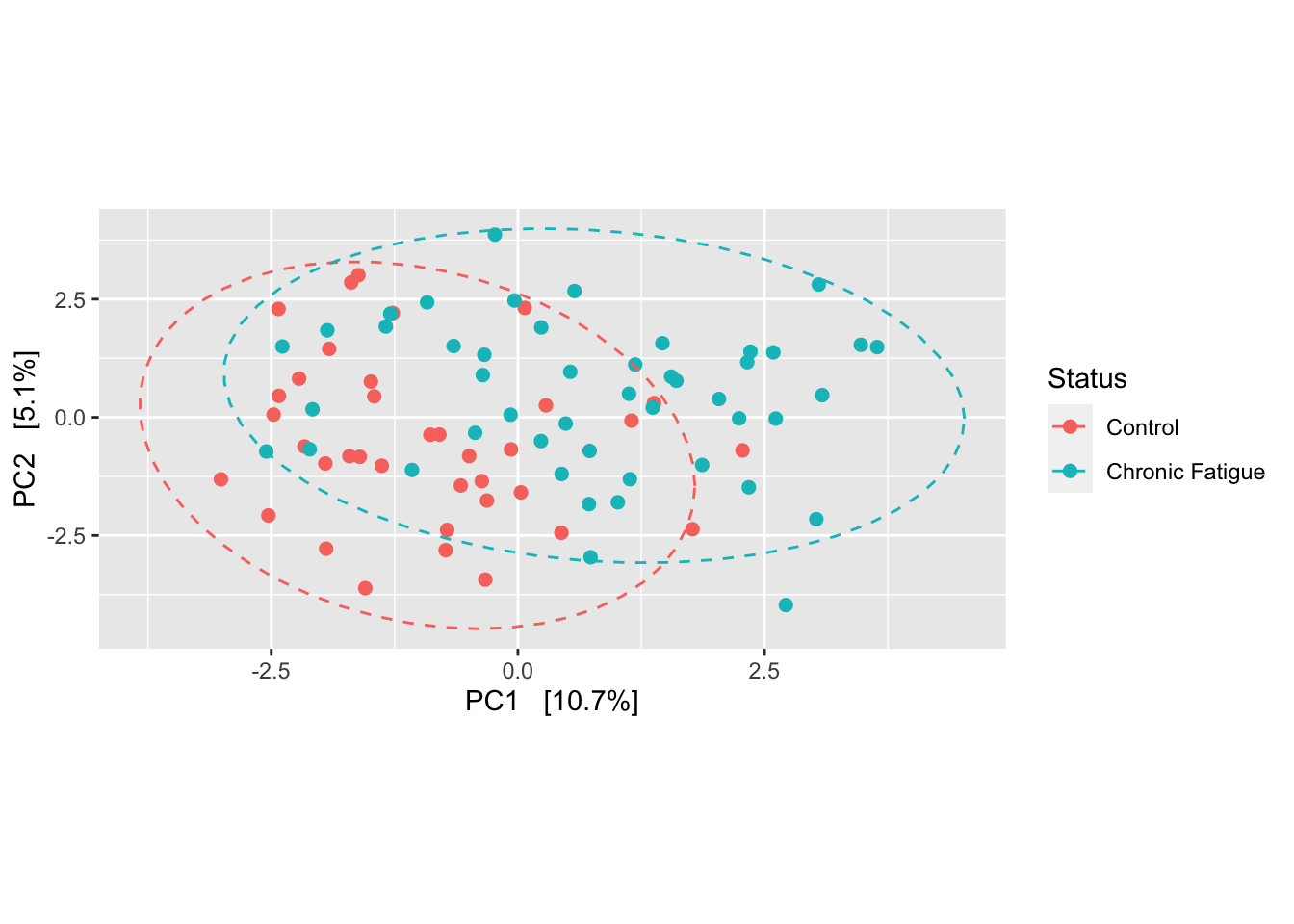
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
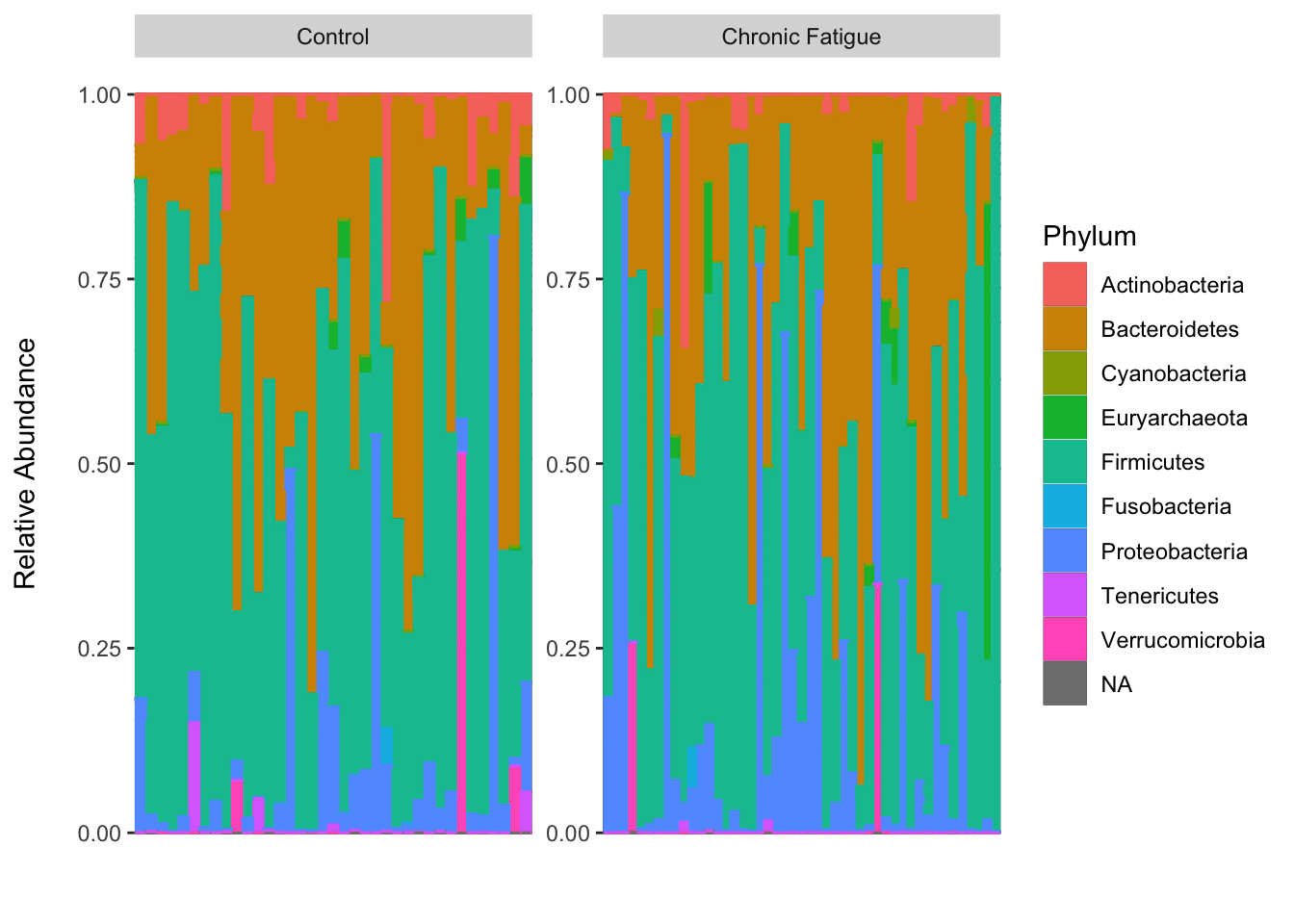
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
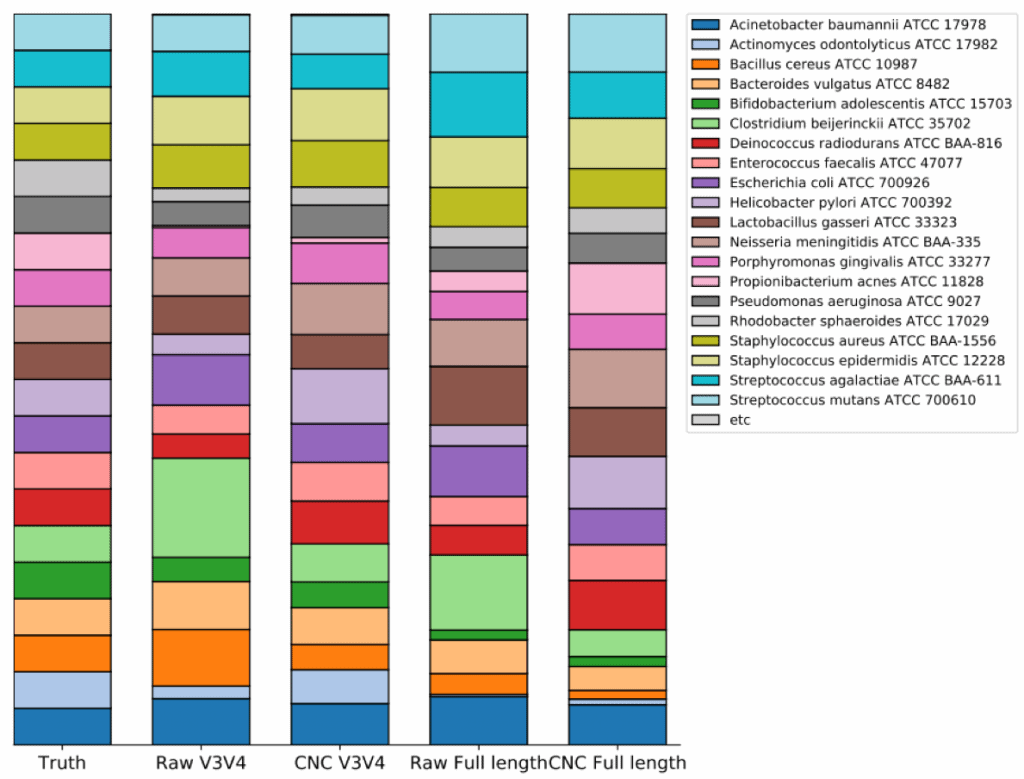
Tutorial 16s Microbiome Analysis Of Atcc R Microbiome Standards Ezbiocloud Help Center

Example Outputs From Microbiomeanalyst A A Box Plot Summary Of The Download Scientific Diagram
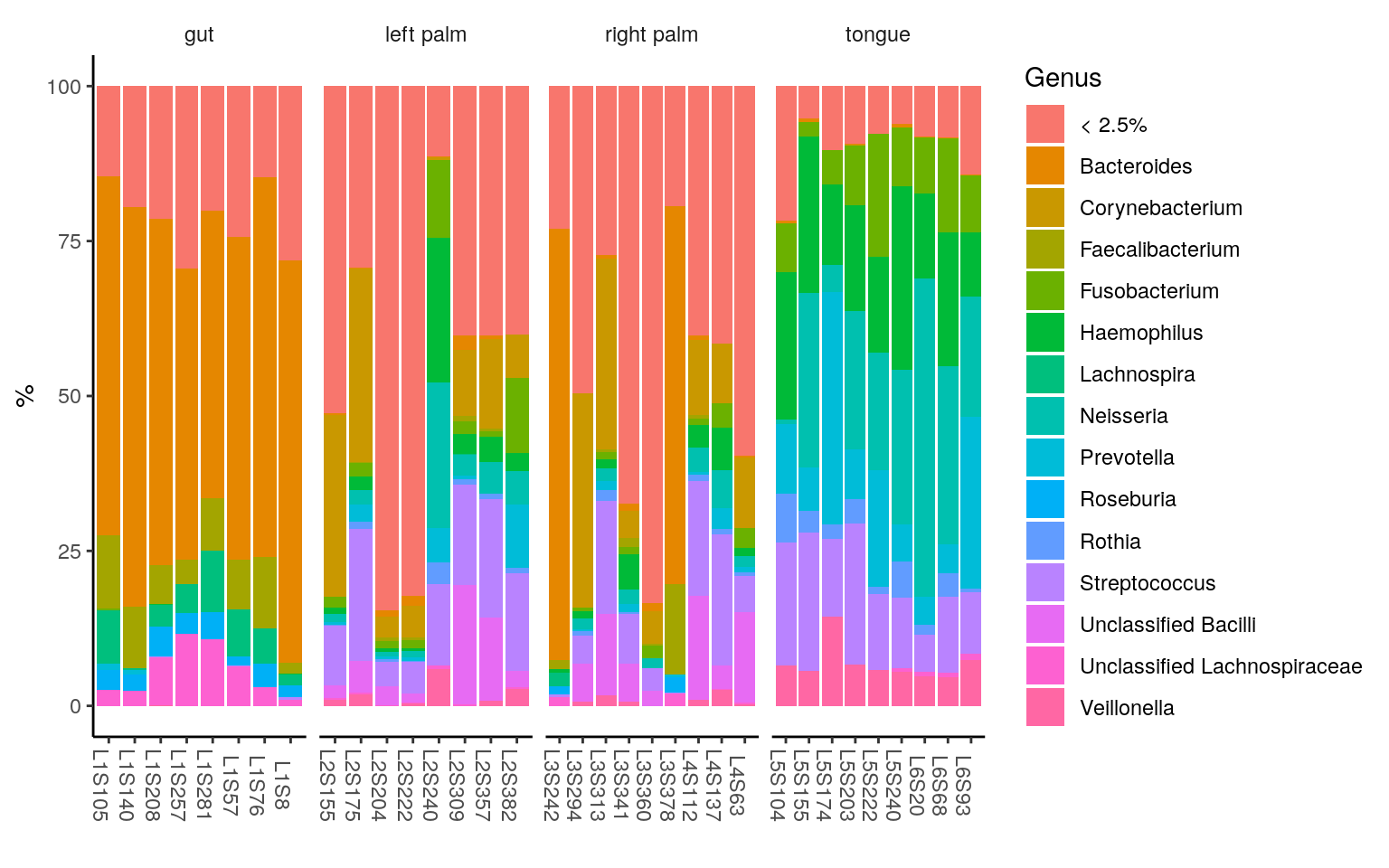
0 komentar
Posting Komentar